A machine learning model can be defined as an output of a rigorous training process and presented as the mathematical representation of a real-world process. A machine learning model is one that is trained to recognize and detect certain patterns present in a dataset. The created model is trained over a set of data, providing it with an algorithm that it can use to reason over and learn from the provided data. Once the model is properly trained, it can be used to reason over data that the model has not seen before and make predictions about that data. Suppose you want to build an application that can recognize a user’s emotions based on their facial expressions. In that case, a model can be easily trained by providing it with images of faces that are each tagged with a certain kind of emotion. Then that trained model can be included in an application that can recognize any user’s emotion. Every machine learning model is categorized in two ways, either supervised or unsupervised. A supervised model can then be further sub-categorized as either a regression or classification model.
The machine learning algorithms help in finding the pattern in a training dataset, which is then used to approximate the target function. In addition, algorithms are responsible for mapping the inputs to the outputs from the available dataset or the dataset being processed. In terms of machine learning, an algorithm can be defined as a run on data to create a machine learning model. Early stages of machine learning, also known as ML for short, saw experiments that only involved theories of computers recognizing patterns from data and learning from them. Today, after building and advancements from those foundational experiments, machine learning is becoming more complex. While machine learning algorithms have been around for a long time, the ability and possibilities to apply them to complex big data applications have been changing more rapidly and effectively with recent developments. Putting them in an application perspective while maintaining a degree of sophistication can set an organization way ahead of its competitors.
Machine learning algorithms also possess the ability to perform pattern recognition where the algorithms learn from data or are fit on a dataset. Many machine learning algorithms have been developed like algorithms for classification, such as k-nearest neighbors or algorithms for regression, such as linear regression or clustering just like k-means. Machine learning algorithms can be described and defined in a model using math and pseudocode. The efficiency and accuracy of both the algorithm and the model can also be analyzed and calculated. Machine learning algorithms can be implemented with any one of a range of modern programming languages such as Python or R. These languages provide a wide range of libraries that can be used to create complex and layered algorithms that the practitioners can use on their projects being developed.
What Is PaDELPy?
PaDELPy is an open-source library that provides a Python wrapper for the PaDEL-Descriptor and a molecular descriptor calculation software. The descriptor can be defined as a mathematical logic that describes the properties of a molecule based on the correlation between the structure of the compound and its biological activity. The PaDEL-Descriptor can be used to work on scientific data to help calculate the molecular fingerprint of specific molecules used to build scientific machine learning models. The PaDEL-Descriptor is a java software that requires running a java file first to help execute and create scientific models. But using the PaDELPy library makes it easier to calculate molecular fingerprints using Python. There is no need to run the jar file, hence reducing the lengthy installation process and aiding quick implementation.
Getting Started with the Code
In this article, we will try to create and implement a scientific machine learning model using the PaDELPy library to calculate the molecular fingerprint and using Random Forest we will be predicting the molecular activity of the drug from the HCV Drug dataset. The following code is inspired by the creators of the PaDELPy library, whose Github repository can be accessed from the link here. If you want to download the raw dataset used in this implementation, you can use the following link.
Let’s Get Started!
Installing the Library
Our first step here will be to install the PaDELPy library. To do so, you can run the following line of code,
#installing the library !pip install padelpy
Loading the PaDELPy Files
Now we will load and properly set up our calculator model first using the necessary PaDELPy files available only in the form of an XML format.
#Downloading the XML data files
!wget https://github.com/dataprofessor/padel/raw/main/fingerprints_xml.zip
!unzip fingerprints_xml.zip
#listing and sorting the downloaded files
import glob
xml_files = glob.glob("*.xml")
xml_files.sort()
xml_files
Output :
['AtomPairs2DFingerprintCount.xml', 'AtomPairs2DFingerprinter.xml', 'EStateFingerprinter.xml', 'ExtendedFingerprinter.xml', 'Fingerprinter.xml', 'GraphOnlyFingerprinter.xml', 'KlekotaRothFingerprintCount.xml', 'KlekotaRothFingerprinter.xml', 'MACCSFingerprinter.xml', 'PubchemFingerprinter.xml', 'SubstructureFingerprintCount.xml', 'SubstructureFingerprinter.xml']
#Creating a list of present files FP_list = ['AtomPairs2DCount', 'AtomPairs2D', 'EState', 'CDKextended', 'CDK', 'CDKgraphonly', 'KlekotaRothCount', 'KlekotaRoth', 'MACCS', 'PubChem', 'SubstructureCount', 'Substructure']
Now we will create a data dictionary with all the loaded and available data files so that we get a key-value pair,
#Creating Data Dictionary fp = dict(zip(FP_list, xml_files)) fp
Output :
{'AtomPairs2D': 'AtomPairs2DFingerprinter.xml',
'AtomPairs2DCount': 'AtomPairs2DFingerprintCount.xml',
'CDK': 'Fingerprinter.xml',
'CDKextended': 'ExtendedFingerprinter.xml',
'CDKgraphonly': 'GraphOnlyFingerprinter.xml',
'EState': 'EStateFingerprinter.xml',
'KlekotaRoth': 'KlekotaRothFingerprinter.xml',
'KlekotaRothCount': 'KlekotaRothFingerprintCount.xml',
'MACCS': 'MACCSFingerprinter.xml',
'PubChem': 'PubchemFingerprinter.xml',
'Substructure': 'SubstructureFingerprinter.xml',
'SubstructureCount': 'SubstructureFingerprintCount.xml'}
Loading the Dataset
With all the necessary PaDELPy files being set up for calculation, we will next load our dataset to be calculated.
#Loading the dataset
import pandas as pd
df = pd.read_csv('https://raw.githubusercontent.com/dataprofessor/data/master/HCV_NS5B_Curated.csv')
#Loading data head
df.head()

#Loading data tail df.tail(2)
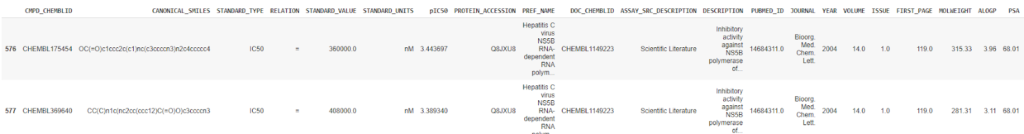
To calculate the molecular descriptor using PaDEL, we will necessarily prepare the data by concatenating the two concerned columns from the dataset, which will act as an input to our model.
#Concatenating necessary columns
df2 = pd.concat( [df['CANONICAL_SMILES'],df['CMPD_CHEMBLID']], axis=1 )
df2.to_csv('molecule.smi', sep='\t', index=False, header=False)
df2

There are 12 fingerprint types present in PaDEL to be calculated from. To calculate all 12, make we will make adjustments to the descriptor types input argument to any of the ones in the fp dictionary variable,
#listing the dictionary pairs
fp
{'AtomPairs2D': 'AtomPairs2DFingerprinter.xml',
'AtomPairs2DCount': 'AtomPairs2DFingerprintCount.xml',
'CDK': 'Fingerprinter.xml',
'CDKextended': 'ExtendedFingerprinter.xml',
'CDKgraphonly': 'GraphOnlyFingerprinter.xml',
'EState': 'EStateFingerprinter.xml',
'KlekotaRoth': 'KlekotaRothFingerprinter.xml',
'KlekotaRothCount': 'KlekotaRothFingerprintCount.xml',
'MACCS': 'MACCSFingerprinter.xml',
'PubChem': 'PubchemFingerprinter.xml',
'Substructure': 'SubstructureFingerprinter.xml',
'SubstructureCount': 'SubstructureFingerprintCount.xml'}
We want to calculate the molecular fingerprint. Hence we will load the necessary file now; we are using the PubChem
#Importing PubChem fp['PubChem']
Setting up the module to calculate the molecular fingerprint,
#Setting the fingerprint module
from padelpy import padeldescriptor
fingerprint = 'Substructure'
fingerprint_output_file = ''.join([fingerprint,'.csv']) #Substructure.csv
fingerprint_descriptortypes = fp[fingerprint]
padeldescriptor(mol_dir='molecule.smi',
d_file=fingerprint_output_file, #'Substructure.csv'
#descriptortypes='SubstructureFingerprint.xml',
descriptortypes= fingerprint_descriptortypes,
detectaromaticity=True,
standardizenitro=True,
standardizetautomers=True,
threads=2,
removesalt=True,
log=True,
fingerprints=True)
Display the calculated fingerprints,
descriptors = pd.read_csv(fingerprint_output_file) descriptors
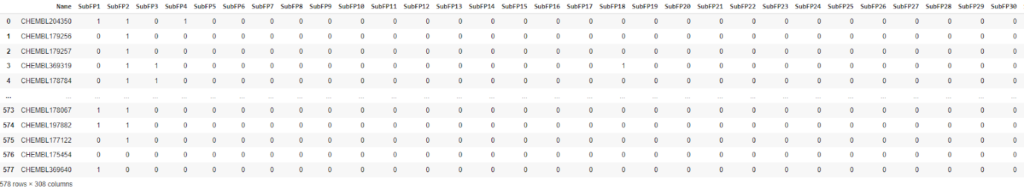
Now we will try to create a classification model from the processed data and Random Forest,
X = descriptors.drop('Name', axis=1)
y = df['Activity'] #feature being predicted
#removing the low variance features
from sklearn.feature_selection import VarianceThreshold
def remove_low_variance(input_data, threshold=0.1):
selection = VarianceThreshold(threshold)
selection.fit(input_data)
return input_data[input_data.columns[selection.get_support(indices=True)]]
X = remove_low_variance(X, threshold=0.1)
X

As we can notice, the data looks pretty much sorted and tells us the most effective drugs. So with this, let’s make predictions from this processed data using our model.
#Splitting into Train And Test from sklearn.model_selection import train_test_split X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2, random_state=42) #Printing Shape X_train.shape, X_test.shape ((462, 18), (116, 18)) #Implementing Random Forest from sklearn.ensemble import RandomForestClassifier from sklearn.metrics import matthews_corrcoef model = RandomForestClassifier(n_estimators=500, random_state=42) model.fit(X_train, y_train)
Output :
RandomForestClassifier(bootstrap=True, ccp_alpha=0.0, class_weight=None,
criterion='gini', max_depth=None, max_features='auto',
max_leaf_nodes=None, max_samples=None,
min_impurity_decrease=0.0, min_impurity_split=None,
min_samples_leaf=1, min_samples_split=2,
min_weight_fraction_leaf=0.0, n_estimators=500,
n_jobs=None, oob_score=False, random_state=42, verbose=0,
warm_start=False)
Making predictions on the drug molecular activity from the created model,
y_train_pred = model.predict(X_train) y_test_pred = model.predict(X_test)
Calculating performance metrics of train split using matthews correlation coefficient,
mcc_train = matthews_corrcoef(y_train, y_train_pred) mcc_train 0.833162800019916
Calculating performance metrics of test split using matthews correlation coefficient,
mcc_test = matthews_corrcoef(y_test, y_test_pred) mcc_test 0.5580628933757674 #performing cross validation from sklearn.model_selection import cross_val_score rf = RandomForestClassifier(n_estimators=500, random_state=42) cv_scores = cross_val_score(rf, X_train, y_train, cv=5) cv_scores array([0.83870968, 0.80645161, 0.86956522, 0.86956522, 0.81521739]) #calcutating mean from the five fold mcc_cv = cv_scores.mean() mcc_cv 0.8399018232819074 #implementing metric test in a single dataframe model_name = pd.Series(['Random forest'], name='Name') mcc_train_series = pd.Series(mcc_train, name='MCC_train') mcc_cv_series = pd.Series(mcc_cv, name='MCC_cv') mcc_test_series = pd.Series(mcc_test, name='MCC_test') performance_metrics = pd.concat([model_name, mcc_train_series, mcc_cv_series, mcc_test_series], axis=1) performance_metrics

As observed from the performance metrics of Random Forest Model created for predicting the molecular drug activity, It seems to perform well on the preset dataset. You can use other algorithms as well to test the performance!
EndNotes
In this article, we understood what exactly a machine learning model stands for and its essence. We also implemented a scientific learning model using the PaDELPy library to calculate the molecular fingerprint using the PaDEL Descriptor and tested the model performance metrics. The following implementation can be found as a colab notebook, which can be accessed using the following link here.