Anomaly Detection techniques have been widely used in data science and now with the rapid increase in temporal data, there has been a huge surge of researchers who are developing new algorithms dealing with outliers across this domain. The time series anomaly detection concentrates to isolate anomalous subsequences of varied lengths. Various statistical methods, supervised and unsupervised methods have already been developed and deep learning methods are the ones which stand out the most as they are extremely capable of dealing with non-linearity and have good learning capacity. But one of the major drawbacks of it is that they have extraordinary potential to fit the data including the outliers.
On the other side, Generative Adversarial Network(GAN) are the generating models, they are known for fully capturing the data’s hidden distribution, but they are not successful learners. Recently, a group of researchers from MIT came up with an idea of Time Series Anomaly Detection using Generative Adversarial Networks(TadGAN)- combining deep learning based approaches and GAN approaches together and developed a benchmarking system for Time Series Anomaly Detection. You can read more about the algorithmic part, here.
In this analysis, we are going to discuss practical implementation of TadGAN through Orion. Orion is a machine learning python-based library for unsupervised time series anomaly detection. This toolkit provides various verified pipelines known as Orion pipelines and uses various AutoML tools developed under DATA TO AI at MIT. Lets dig in!
Installation
Easiest way to install is through pip.
# install dependencies then restart kernel and run again
! pip install orion-ml
! pip install 'urllib3!=1.25.0,!=1.25.1,<1.26,>=1.21.1'
! git clone https://github.com/signals-dev/Orion.git
#moving all the required modules in current working directory
! mv Orion/notebooks/tulog/* .Importing all the required modules and functions
# general imports
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
#importing sklearn module
from sklearn.impute import SimpleImputer
from sklearn.preprocessing import MinMaxScaler
#utils.py contains all the plot function.
from utils import plot, plot_ts, plot_rws, plot_error, unroll_tsDataset
The dataset which will be taking, is NYC taxi dataset. This dataset is maintained by the Numenta community. The full raw version is maintained by The New York City Taxi and Limousine Commission (TLC). The processed dataset contains the demand over a period of 7 months recorded in every 30 minutes.
Importing the dataset. There are many datasets which are already available in Orion.
#importing data module to load the dataset
from orion.data import load_signal, load_anomalies
#Import nyc_taxi dataset
signal = 'nyc_taxi'
# load_signal function load the given dataset
df = load_signal(signal)
# Since this dataset is already labelled, we will use load_anomalies functions to get all the known anomalies.
known_anomalies = load_anomalies(signal)
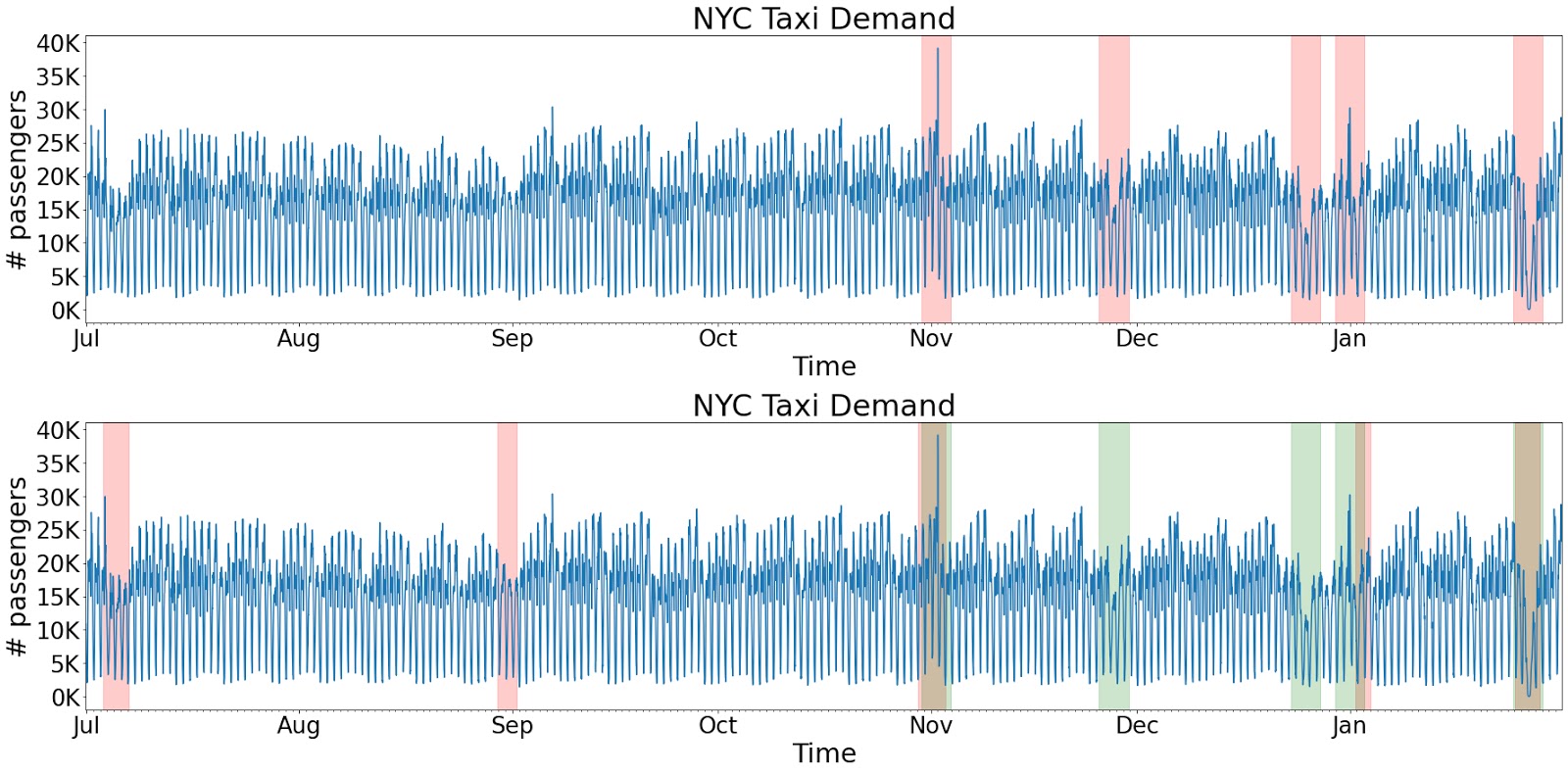
df.head(5)Plotting the dataset with the known anomalies. The blue color represents all the data points and the light pink colored patch represents the known anomaly.
#the pink-colored patch represent known anomaly
plot(df, known_anomalies)ORION API
Through this API, we will be doing anomaly detection, using TadGAN model. As stated earlier, there are many pipelines like ARIMA, LSTM, etc, available in Orion, you can use any of them. Our main focus will be TadGAN pipeline. The procedure to use this library is easy as scikit-learn. At first, we train the data using the fit method and to do anomaly detection, we use the detect method. In this case, we will be using the fit_detect method. This process might take some time. Once it is over, it can be easily visualized and results can be seen.
Note: For experimentation purposes, you can reduce the number of epochs in the tadgan.json file such that you reduce the number of training iterations.
#importing Orion pipeline
from orion import Orion
#loading the TadGAN pipeline
orion = Orion(
pipeline='tadgan.json')
#fitting the data to the TadGAN pipeline of Orion and detecting the outliers.
anomalies = orion.fit_detect(df)Let’s plot the result.
plot(df, [anomalies, known_anomalies])
anomalies.head(5)In the plot above, the red intervals represent the detected anomalies, the green intervals show the known anomalies. Hence, the model was able to detect some anomalies different from known anomalies(ground truth).
Note: the results might differ between runs.
Trace Back to functioning of Model
We have directly passed tadgan.json into the Orion pipeline and abracadabra, we got the results directly. This process is not easy as it seems. If we carefully look into tadgan.json file, we will get to know that it is actually doing multiple processing like data-preprocessing, model training, to post-processing functionalities. As defined by the author, these functions inside this file are called primitives. Each primitive is responsible for a specific task which we will be looking further into this article.
Data Preparation
- Data Frequency
This is responsible for adjusting spacing. Two important parameters in this are:
Interval: An integer of time span, to compute the aggregation. In this example, we have taken interval=1800, which is equivalent to 30 minutes. Previously, we stated that each data point is taken over a period of 30 minutes that means it is equally spaced. Hence, in our case we can skip this preprocessing step.
Method: This represents the type of aggregation method to use like mean, mode or median. Default is also mean.
def time_segments_aggregate(X, interval, time_column, method=['mean']):
"""Aggregate values over given time span.
Args:
X (ndarray or pandas.DataFrame):
N-dimensional sequence of values.
interval (int):
Integer denoting time span to compute aggregation of.
time_column (int):
Column of X that contains time values.
method (str or list):
Optional. String describing aggregation method or list of strings describing multiple
aggregation methods. If not given, `mean` is used.
Returns:
ndarray, ndarray:
* Sequence of aggregated values, one column for each aggregation method.
* Sequence of index values (first index of each aggregated segment).
"""
#checking for the input datatype as numpy array and converting it to dataframe
if isinstance(X, np.ndarray):
X = pd.DataFrame(X)
#sorting the values on timestamp column and setting it as a index
X = X.sort_values(time_column).set_index(time_column)
if isinstance(method, str):
method = [method]
start_ts = X.index.values[0]
max_ts = X.index.values[-1]
values = list()
index = list()
while start_ts <= max_ts:
end_ts = start_ts + interval
subset = X.loc[start_ts:end_ts - 1]
aggregated = [
getattr(subset, agg)(skipna=True).values
for agg in method
]
values.append(np.concatenate(aggregated))
index.append(start_ts)
start_ts = end_ts
return np.asarray(values), np.asarray(index)
#here df is the given dataframe and "timestamp" is the required column to be altered.
X, index = time_segments_aggregate(df, interval=1800, time_column='timestamp')- Data Imputation
#Using the simple scikit imputer
imp = SimpleImputer()
X = imp.fit_transform(X)- Data Normalization
Normalize the data into the range of [-1,1] using MinMaxScalar of scikit-learn library.
#Normalizing the data using scikit-learn MinMaxScalar
scaler = MinMaxScaler(feature_range=(-1, 1))
X = scaler.fit_transform(X)- Data Slicing
This method divides the original time series into signal segments as shown below.
def rolling_window_sequences(X, index, window_size, target_size, step_size, target_column,
drop=None, drop_windows=False):
"""Create rolling window sequences out of time series data.
The function creates an array of input sequences and an array of target sequences by rolling
over the input sequence with a specified window.
Optionally, certain values can be dropped from the sequences.
Args:
X (ndarray):
N-dimensional sequence to iterate over.
index (ndarray):
Array containing the index values of X.
window_size (int):
Length of the input sequences.
target_size (int):
Length of the target sequences.
step_size (int):
Indicating the number of steps to move the window forward each round.
target_column (int):
Indicating which column of X is the target.
drop (ndarray or None or str or float or bool):
Optional. Array of boolean values indicating which values of X are invalid, or value
indicating which value should be dropped. If not given, `None` is used.
drop_windows (bool):
Optional. Indicates whether the dropping functionality should be enabled. If not
given, `False` is used.
Returns:
ndarray, ndarray, ndarray, ndarray:
* input sequences.
* target sequences.
* first index value of each input sequence.
* first index value of each target sequence.
"""
out_X = list()
out_y = list()
X_index = list()
y_index = list()
target = X[:, target_column]
if drop_windows:
if hasattr(drop, '__len__') and (not isinstance(drop, str)):
if len(drop) != len(X):
raise Exception('Arrays `drop` and `X` must be of the same length.')
else:
if isinstance(drop, float) and np.isnan(drop):
drop = np.isnan(X)
else:
drop = X == drop
start = 0
max_start = len(X) - window_size - target_size + 1
while start < max_start:
end = start + window_size
if drop_windows:
drop_window = drop[start:end + target_size]
to_drop = np.where(drop_window)[0]
if to_drop.size:
start += to_drop[-1] + 1
continue
out_X.append(X[start:end])
out_y.append(target[end:end + target_size])
X_index.append(index[start])
y_index.append(index[end])
start = start + step_size
return np.asarray(out_X), np.asarray(out_y), np.asarray(X_index), np.asarray(y_index)
#the target value; the value at time t.
#previous observed values, this is determined by the window width.
X, y, X_index, y_index = rolling_window_sequences(X, index,
window_size=100,
target_size=1,
step_size=1,
target_column=0)Window slicing done by above function can be seen as
#function from utils.py module. Representing all the windows that has been created by slicing
#Here X represents the input used to train the model. In the previous example, we see X has 10222 training data points.
#Notice that 100 represents the window size. On the other hand, y is the real signal after processing,
#which we will use later on to calculate the error between the reconstructed and real signal.
plot_rws(X)Training and Detection
As stated in the Research Paper, the architecture of this model requires four neural networks.
- encoder: maps X to its latent representation Z.
- generator: maps the latent variable Z back to X, which we will denote later on as X_hat.
- criticX: distinguishes between the real time series sequences from X and generated time series from generator(Z) or X_hat.
- criticZ: measures the performance of the mapping into latent space or in simpler words, it discriminates between Z and encoder(X).
More details of the composition of each network is mentioned in model.py. Specifying all the parameters below, fitting the model to the input X.
from model import hyperparameters
from orion.primitives.tadgan import TadGAN
hyperparameters["epochs"] = 35
hyperparameters["shape"] = (100, 1) # based on the window size
hyperparameters["optimizer"] = "keras.optimizers.Adam"
hyperparameters["learning_rate"] = 0.0005
hyperparameters["latent_dim"] = 20
hyperparameters["batch_size"] = 64
tgan = TadGAN(**hyperparameters)
tgan.fit(X)Through this image, it is clear that TadGAN is almost able to reconstruct the actual data. Now, the difference between the original data and reconstructed data will be used to calculate the loss(error).
Plot of original and reconstructed signal
Error Computation
In the TadGAN pipeline, we use tadgan.score_anomalies to perform error calculation. It is a smoothed error function that uses a window based method to smooth the curve then uses either: area, point difference, or dtw as a measure of discrepancy.
- Area: It captures the general shape of actual and reconstructed signals and then compares them.
- Point: It compares each point between the original and reconstructed signal. It is considered as a strict method and does not allow any mistakes.
- Dynamic Time Warping (DTW): It compares two signals together using any pairwise distance measure but it allows for one signal to be lagging behind another. A lenient method.
Plotting the error
from orion.primitives.tadgan import score_anomalies
error, true_index, true, pred = score_anomalies(X, X_hat, critic, X_index, rec_error_type="dtw", comb="mult")
pred = np.array(pred).mean(axis=2)
# visualize the error curve
plot_error([[true, pred], error])From the above graph, we can see that high peaks represent high error values; now we just have to classify them as an anomalous point by giving a threshold value.
# threshold to classify the high peak data points as anomolous points
thresh = 10
intervals = list()
i = 0
max_start = len(error)
while i < max_start:
j = i
start = index[i]
while error[i] > thresh:
i += 1
end = index[i]
if start != end:
intervals.append((start, end, np.mean(error[j: i+1])))
i += 1
intervalsNow we will plot the actual data with known anomalies and predicted anomalies. The red intervals represent the detected anomalies, the green intervals show the known anomalies. If we check the plot carefully, then we will come to know it has missed two or three anomalies and that is because of the thresholding method. Hence we will incorporate those missed-out anomalies by using window based methods.
In this method, we initially define the window size we want to analyze and then find the anomalous signals by looking at the mean and standard deviation in that window and then repeat this procedure by saving all the start and stop indexes and moving to the next window and at last, we combine them.
from orion.primitives.timeseries_anomalies import find_anomalies
# find anomalies
intervals = find_anomalies(error, index,
window_size_portion=0.33,
window_step_size_portion=0.1,
fixed_threshold=True)
intervalsPlotting again the actual data with known anomalies and predicted anomalies.
You can see, this plot is the same as we got earlier from Orion API. Hurray!
End-to-End Pipeline Configuration
For configuration of the pipeline, we adjust the parameters in tadgan.json or by passing the dictionary of parameters. The parameters mentioned in the dictionary will be overridden. Here, I changed the number of epochs to 15 and fit the TadGAN again to the dataset. For more information, you can check the documentation.
from orion import Orion
#paramter dictionary contains the parameter which are to be overridden.
parameters = {
"mlprimitives.custom.timeseries_preprocessing.time_segments_aggregate#1": {
"interval": 3600 # hour level
},
'orion.primitives.tadgan.TadGAN#1': {
'epochs': 15,
}
}
orion = Orion(
'tadgan.json',
parameters
)
anomalies = orion.fit_detect(df)Now again plotting the anomalies.
In the plot we can see that the result is quite different from what we got earlier. It can be because of the parameter we tweaked like number of epochs, interval or it can be because the model is not trained enough. We can check all of this with the help of vizualization parameter in the detect to return the intermediate output, in which we are interested. With the help of visualization parameter in tadgan.json, we can return the following variables.
- X: this is the output of the preprocessing steps from averaging, imputing, and scaling. These steps were showcased previously as steps (A, B, and C).
- X_hat: this is the “predicted” output by the TadGAN model without any processing. It represents the reconstructed window at each timepoint.
- es: this is the error calculated by capturing the discrepancies between original and reconstructed signal.
then we can use anomalies, viz = orion.detect(df, visualization=True) where viz will be a dictionary of intermediate outputs.
Before discussing the methods, let us take an example. For this, create a dummy dataset containing three ground truth values and two anomalies.The code for creating and plotting the dummy dataset is available here. Now, there are two methods for evaluation:
Weighted Segment: It assesses each datapoint in the detected anomalies with its counterpart in the ground truth. It separates the signal into partitions based on the ground truth and anomaly and then it compares each segment to another and creates a confusion matrix. The overall score is weighted by the duration of each segment. Here is an example of how we can use the weight segment in orion. For more info, check the documentation linked in the heading.
#we can use orion.evaluation subpackage to compute multiple metrics using the weighted segment approach.
#For example to compute the accuracy, we use contextual_accuracy(..., weighted=True).
#There are other metrics available, for reference checkout the orion.evaluation documentation.
from orion.evaluation.contextual import contextual_accuracy, contextual_f1_score
accuracy = contextual_accuracy(ground_truth, anomalies, start=start, end=end)
f1_score = contextual_f1_score(ground_truth, anomalies, start=start, end=end)
print("Accuracy score = {:0.3f}".format(accuracy))
print("F1 score = {:0.3f}".format(f1_score))Overlapping Segment: It assesses the detected anomaly segment by checking if it overlaps with the correct anomaly. It records following values:
1) TP, if a ground truth segment overlaps with the detected segment.
2) FN, If the ground truth segment does not overlap any detected segments.
3) FP, If a detected segment does not overlap any labeled anomalous region.
#Similarly, we can use the same metric functions, but with parameter weighted=False.
#Note: overlap segment approach, does not account for true negatives.
#Reason being, anomalies in time series data are rare and so "normal" instances will skew the value of the computed metric.
#Therefore, using this approach we cannot compute metrics such as the accuracy.
f1_score = contextual_f1_score(ground_truth, anomalies, start=start, end=end, weighted=False)
print("F1 score = {:0.3f}".format(f1_score))Pipeline Evaluation End-to-End
Orion API has an option to evaluate the whole pipeline(with its known labels). Most of the part of its code has already been explained above in Orion API. So, the main focus will be on the evaluation here. A list of metrics is used to evaluate the whole pipeline. Here, metrics list contains all the function names which compares the gound truth with detected anomaly and returns a metric value. Some of the predefined metrics in Orion, such as:
- Accuracy
- F1 Score
- Precision
- Recall
By default, we use the weighted segment approach, you can override metrics defined by specifying it in a new metrics dictionary.
from orion import Orion
from orion.data import load_signal, load_anomalies
#Importing all the methods for evaluation
from orion.evaluation.contextual import contextual_accuracy, contextual_f1_score, contextual_precision
metrics = [
'f1',
'recall',
'precision',
]
orion = Orion(
'tadgan.json'
)
signal = 'nyc_taxi'
# load signal
df = load_signal(signal)
# load ground truth anomalies
ground_truth = load_anomalies(signal)
#Evaluation scores mentioned in the metrics list
scores = orion.evaluate(df, ground_truth, fit=True, metrics=metrics)
scoresConclusion
In the above Tutorial we have discussed the Orion Pipeline for TadGAN from both the API and scratch method, End-to-End Pipeline Configuration and evaluation methods.
Tutorial and other resources used above.