PyTorch recently announced the release of its machine learning drug discovery platform TorchDrug to accelerate drug discovery research. The library is open-sourced and can be installed through pip if you have PyTorch and torch-scatter installed using
pip install torchdrug, or through
conda conda install -c milagraph -c conda-forge torchdrug.
TorchDrug covers many recent techniques such as graph machine learning, deep generative models, and reinforcement learning. It also provides reusable training and evaluation routines for popular drug discovery tasks, including property prediction, pretrained molecular representations, de novo molecule design, retrosynthesis and biomedical knowledge graph reasoning. It is easy to build a prototype for one’s own dataset and application based on these techniques and modules.
For advanced users, the platform provides multiple levels of building blocks for different customisation demands. These include low-level data structures and operations (e.g. molecules and graph masking), mid-level layers and modules (e.g. graph convolutions and GNNs) and high-level task routines (e.g. property prediction). TorchDrug is flexible for all kinds of customisation. It also provides graph data structures and operations for manipulating biomedical objects, as well as reusable layers, models and tasks for building machine learning models.
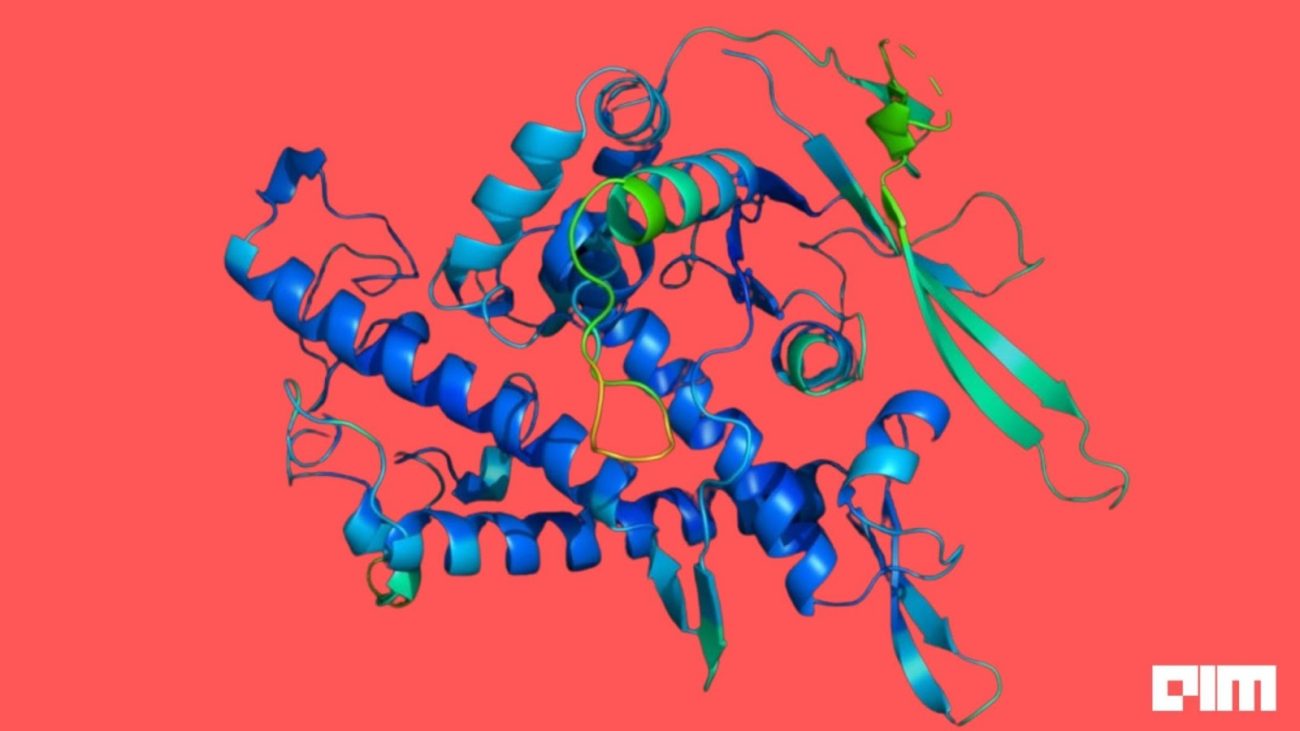
The core data structures of TorchDrug are graphs, which can be used to represent a wide range of biological objects, including molecules, proteins and biomedical knowledge graphs. Visualisation API in the library can be used to check graph objects.
PackedGraph data structure, which builds a unified large graph and re-index each small graph in the batch, can be used to create a batch of variable-size graphs.
Code for calculating a batch of 4 molecules:
mols=data.PackedMolecule.from_smiles(["CCSCCSP(=S)(OC)OC", "CCOC(=O)N", "N(Nc1ccccc1)c2ccccc2", "NC(=O)c1cccnc1"])
mols.visualize()
mols = mols.cuda()
print(mols)
# PackedMolecule(batch_size=4, num_nodes=[12, 6, 14, 9], num_edges=[22, 10, 30, 18], device='cuda:0')
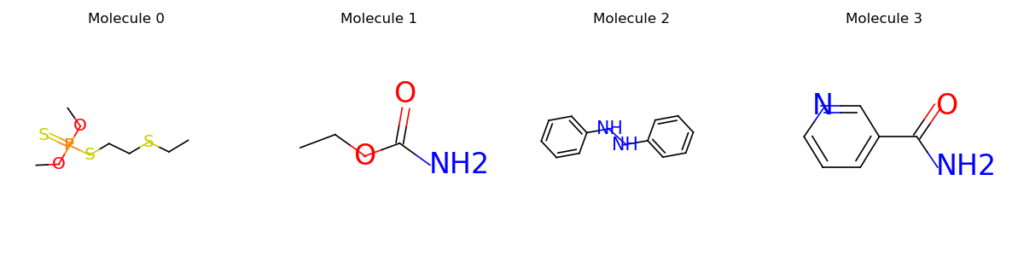
Graphs also support a wide range of indexing operations. Typical usages include applying node masking, edge masking or graph masking. The optimiser can be used for parameters in the task and combine everything into the core. The engine provides convenient routines for training and testing. To test the model on the validation set, it only takes one line.
TorchDrug is designed to cater to all kinds of development. This ranges from low-level data structures and operations, mid-level layers and models, to high-level tasks. One can easily customise modules at any level with minimal effort by utilising building blocks from a lower level.
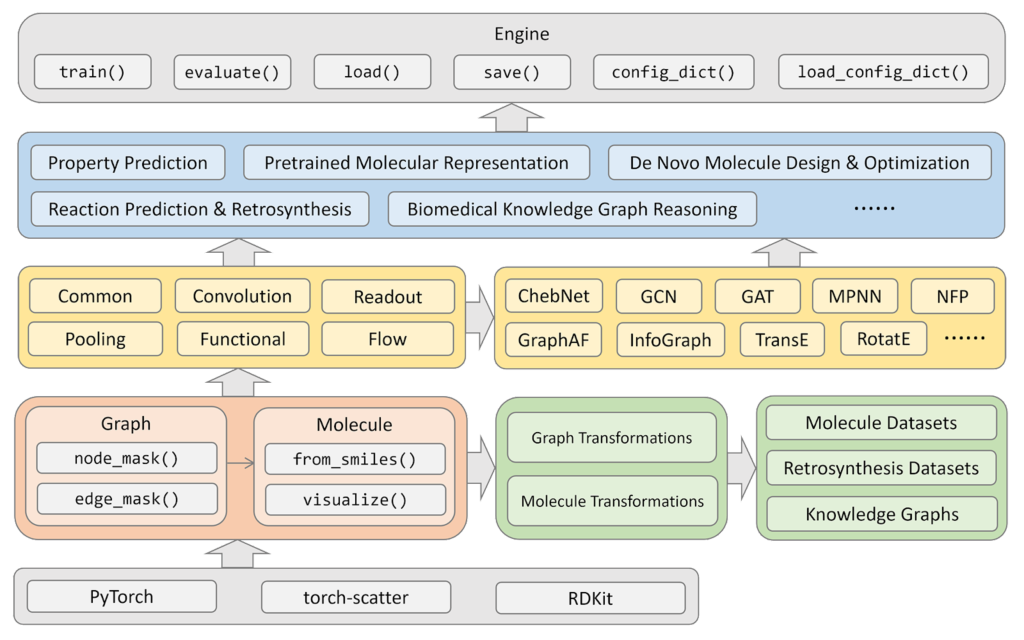
The correspondence between modules and the hierarchical interface is :
- torchdrug.data: Graph data structures and graph operations; e.g. molecules.
- torchdrug.datasets: Datasets; e.g. QM9.
- Torchdrug.layers: Neural network layers and loss layers; e.g. message-passing layer.
- Torchdrug.models: Representation learning models; e.g. message passing neural network.
- torchdrug.tasks: Task-specific routines; e.g. molecule property prediction.
- Torchdrug.core: Engine for training and evaluation.
Machine learning for drug discovery is a fast-growing area, and the PyTorch team expects that TorchDrug could help more and more people get involved in this interdisciplinary area. To learn more about TorchDrug, you can check out the Colab tutorials for basic usage and several drug discovery tasks using the link here.