|
Listen to this story
|
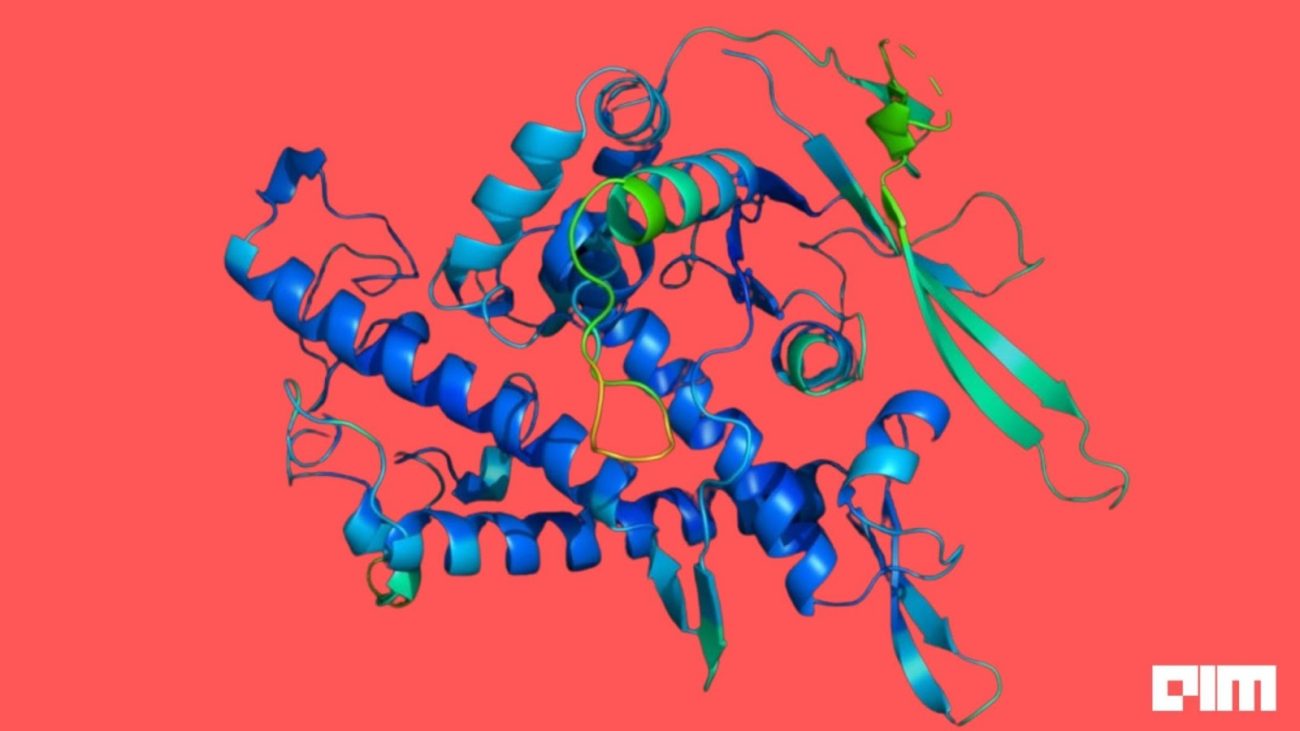
Back in 2020, Google AI’s AI arm DeepMind participated in a protein-structure prediction challenge called CASP or Critical Assessment of Structure Prediction and beat 100 other teams in solving one of the most difficult problems in Biology — figuring out a protein’s 3D shape from its amino acids.
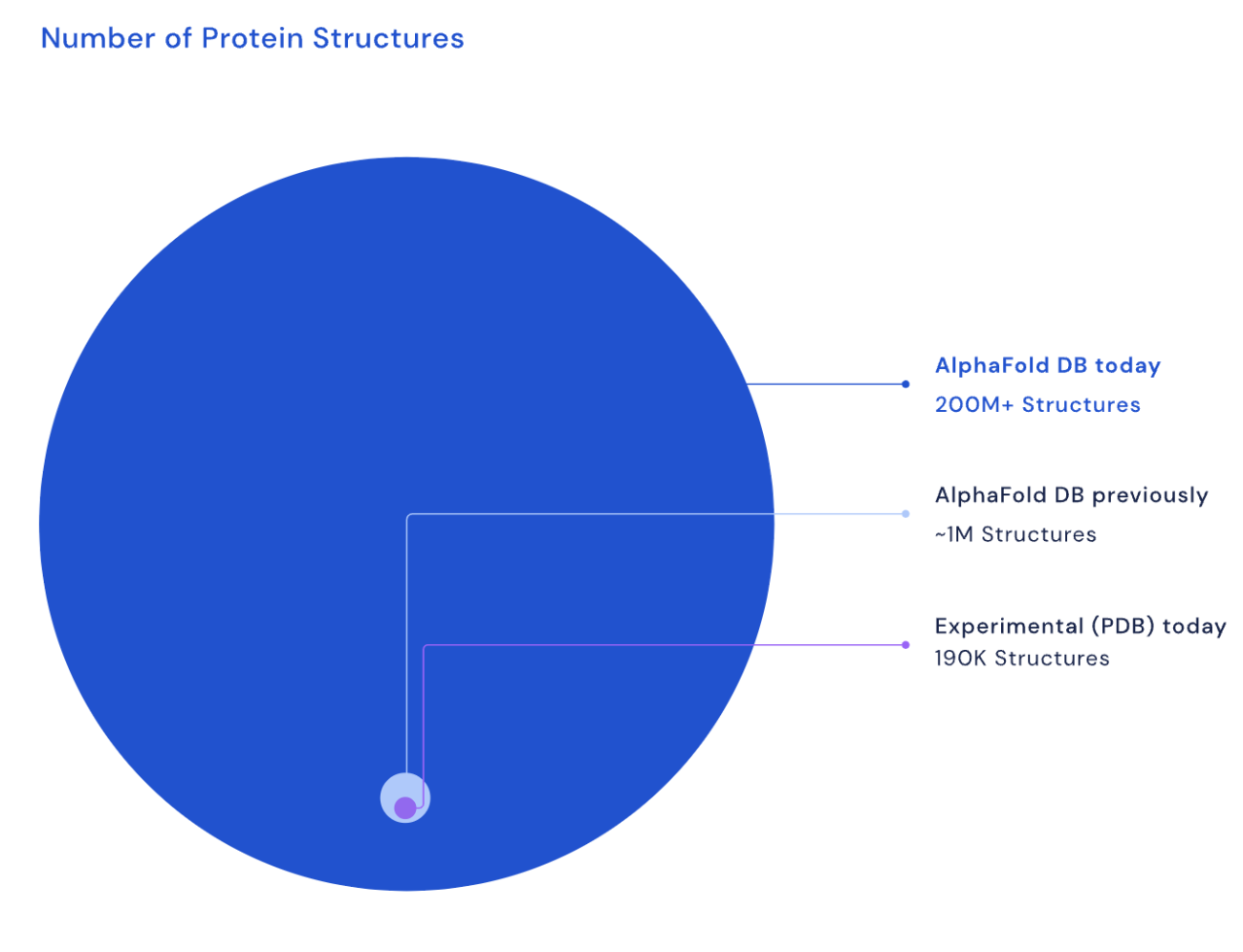
Two years into this breakthrough, DeepMind’s AI software AlphaFold has taken quick leaps in predicting protein structures. Last year, DeepMind started releasing AlphaFold’s predictions using a publicly available database that it built in collaboration with European Molecular Biology Laboratory (EMBL). This initial dataset included 98% of all human proteins. Last month, AlphaFold released an expanded database with more than 200 million protein structures encompassing almost every protein existing in Science.

To most, this feat has turned bystanders who were sceptical about AI’s role in pharma into believers. Protein folding has been a 50-year old problem in Biology. Scientists, since the 1990s, have been trying to train their computers to predict protein structures but largely met with failure. The reward of finding a solution to this grand problem was immense.
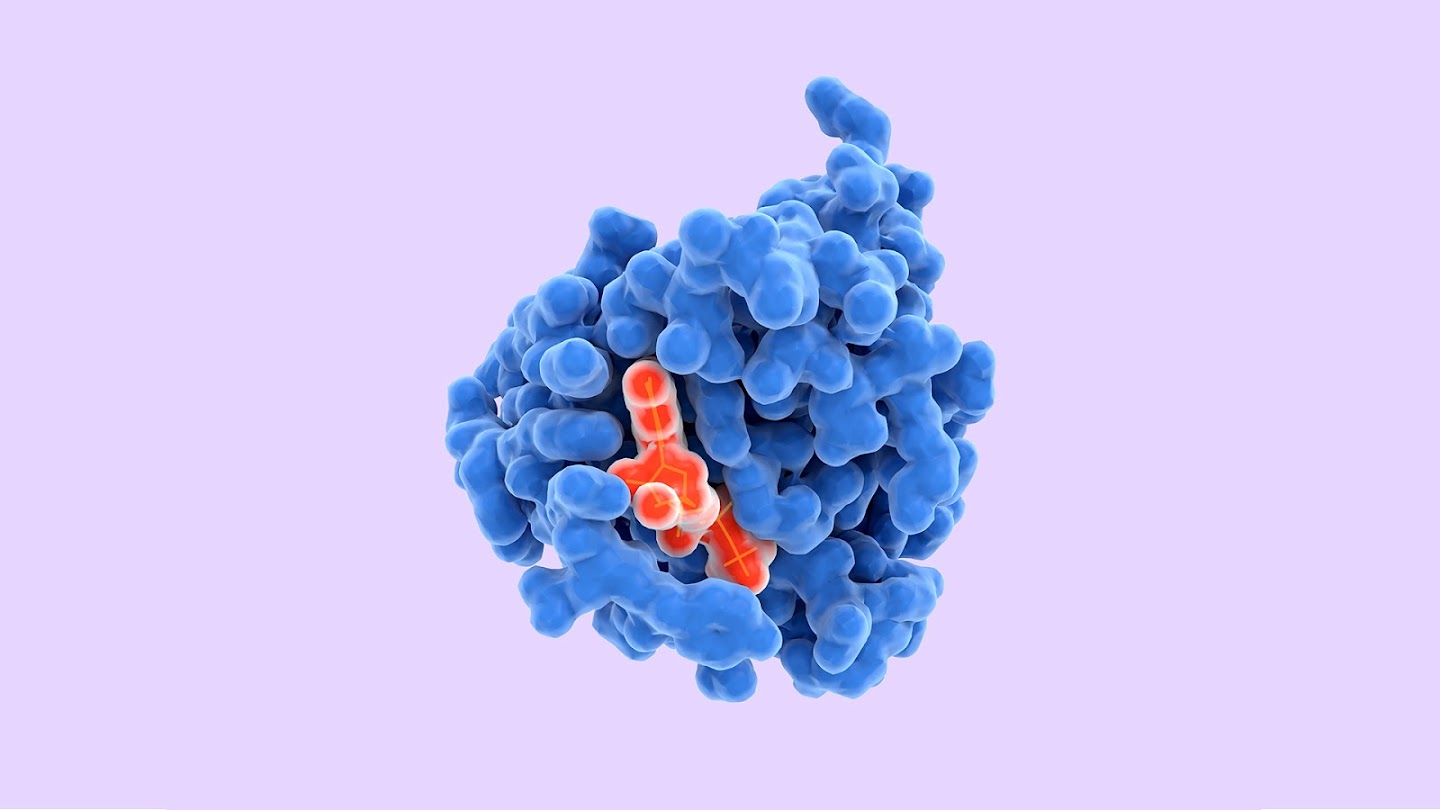
Proteins are the building blocks of life with every structure having a different shape. Understanding how protein structures fold into three-dimensional shapes meant having an insight about the role of proteins and how these macromolecules behaved. Protein misfolding contributes to the development of diseases like Alzheimer’s. AlphaFold’s achievements could mean that scientists will be able to study neurodegenerative diseases and design drugs faster.
Mere AI marvel or really helpful?
All this progress, however, comes with a caveat. Needless to say, AlphaFold’s predictions are a success for pattern recognition and data wrangling. However, what’s worth remembering despite the grand headlines is that the protein structures that AlphaFold has designed are predictions and not the actual protein structures. This makes the designs far less valuable than the actual data of protein structures obtained from X-Rays and NMRs or Nuclear Magnetic Resonance instruments.
On the one hand, the model has created a huge bank of new protein structure predictions that weren’t expected at all, while on the other, AlphaFold’s algorithm fails when it comes to disordered protein regions. An intrinsically disordered protein or IDP doesn’t have a fixed 3D structure, usually during the absence of other macromolecules like RNA. Other protein structures never follow structure under any conditions. More so, this is the main characteristic of proteins that helps them function.
Protein structures innately shift and transform, sometimes very drastically and sometimes in subtle ways when small-molecule ligands show up. It will be difficult for AlphaFold to be able to predict these tweaks considering there are very few small-molecule ligand proteins to train them on. While there are only close to 20 of them found, the number of molecular structures is so large that there are infinite combinations possible.
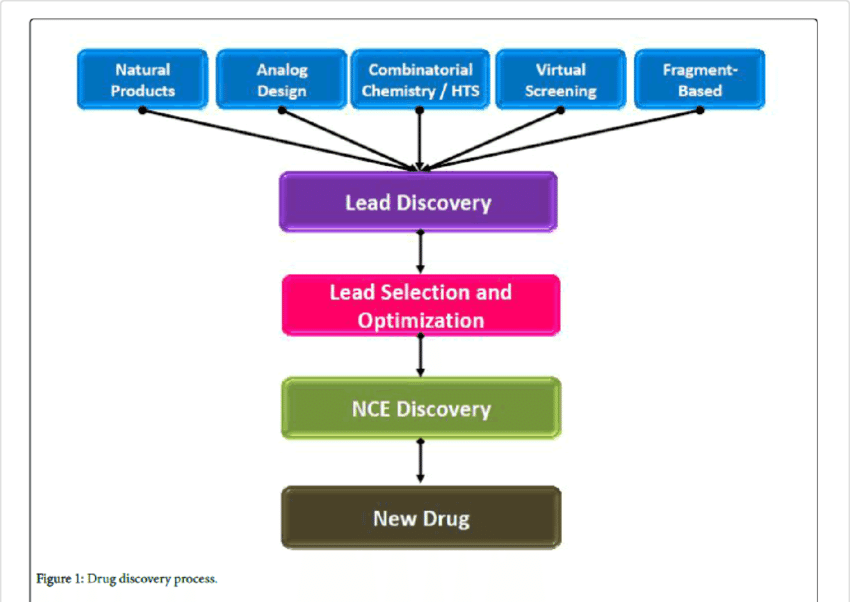
The coverage around AlphaFold may also be overblown in the context of drug discovery since protein structures are almost never a part of the rate-limiting step in the process. Projects revolving around drug discovery normally use living cells for pure protein. Additionally, the context given by protein structures contribute to a very minuscule portion of making a drug. Scientists from Swiss multinational healthcare firm Roche have confirmed this, saying that while this could be helpful, it didn’t solve the whole problem.
Drug discovery is an arduous process where trials are conducted to understand how the compounds are reacting. In the life cycle of a drug, success is signalled by how cells and organs behave in a specific organism when the protein is disturbed. Deep into the process, real data is what helps testing for metrics like metabolism, toxicology and pharmacokinetics where a prediction of a protein can hardly help.

New horizons
On the flipside, AlphaFold’s software learns from low-level structures that it has been exposed to. Despite the fact that these structures may have no precedence, it does offer a base level for making more concrete design predictions.
This lack of analogy is what also helps the case of AlphaFold. These predictions could help the model make de novo proteins, which is a process inverse to the protein folding problem. AlphaFold can help scientists design protein structures from scratch rather than using a known protein. While it may be a while until this happens, the potential lies where experts can computationally predict how the proteins will fold and what their stable condition is. There is a possibility that these properties can be tuned then depending on the chosen application. This could eventually lead to entirely new areas of research in Biology. Admittedly, while this can fasten the pace of research in the earliest stages, there is little scope to quicken the process of drug discovery itself.

In two recent instances, researchers at The University of California, San Francisco, used AlphaFold and cryogenic electron microscopy to study Nsp2, a protein part of the deadly SARS-CoV-2 viruses. AlphaFold was able to determine that the protein has a zinc ion-binding. The protein plays a role in RNA binding, which could open up other areas for research.
Open-source computational approaches like AlphaFold’s are also apt in research areas for diseases that are often neglected. DeepMind has collaborated with the Drugs for Neglected Disease Initiative or DNDi in Geneva, which intends to investigate rare diseases like Chagas, a life-threatening illness caused by the Trypanosoma cruzi parasite. The researchers have found a molecule that can bind itself to a protein in the parasite and kill it. AlphaFold can help with identifying the protein’s structure to treat Chagas disease.