Last week, the Alphabet-owned research lab DeepMind announced that it is making AlphaFold 2.0 source code public. This AI-based algorithm predicts the shape of proteins, a major challenge in the healthcare and life sciences field. With this decision, DeepMind hopes to offer easy access and better research opportunities to the scientific community in areas such as drug discovery.
Last year we presented #AlphaFold v2 which predicts 3D structures of proteins down to atomic accuracy. Today we’re proud to share the methods in @Nature w/open source code. Excited to see the research this enables. More very soon!https://t.co/6uiV51Xly5https://t.co/CLo7EKubBT pic.twitter.com/5dAgg9mOMN
— Demis Hassabis (@demishassabis) July 15, 2021
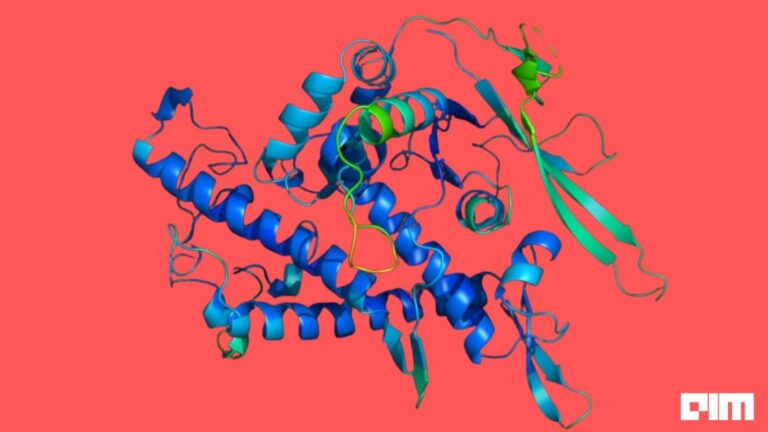
AlphaFold 2.0
Proteins consist of amino acids and are the building blocks of tissues, muscles, hair, antibodies, and other enzymes. They underpin every biological process ever known to us. Each protein has an intricate 3D shape that defines what it does and how it works. However, deducing the structure of proteins is a very tedious, tricky and time-consuming task. Existing methods, such as X-ray crystallography, Nuclear Magnetic Resonance spectroscopy, or cryogenic electron microscopy, cost millions of dollars and employ a laborious trial and error approach.
In 2016, DeepMind began to tackle the protein folding problem by leveraging artificial intelligence. In 2018, AlphaFold 1.0 was released, though it wasn’t good enough to employ researchers in the field. After further research, AlphaFold 2.0 was released in December 2020 and the algorithm has been making waves ever since.
It is an upgrade over the previous version. It is composed of a system of subnetworks integrated into a single differentiable end-to-end model based on pattern recognition and trained to function as an integrated platform.
This algorithm is a neural net, quite similar to the one used for image recognition. The input in the algorithm is the information about protein sequences. First, this model estimates the distance between every single residue in the protein structure. This distance data on every such protein collected is then parsed through a smooth gradient technique, which folds the protein into a structure compatible with the distance measured in the previous step. Finally, the algorithm combines all distance predictions with the Rosetta energy function to redefine the final structure.
Open-sourcing AlphaFold 2.0
From diagnosing fatal diseases to drug discovery, the knowledge of a protein structure will enable us to tackle problems previously thought impossible. Predicting protein structure could be useful in future pandemic response efforts as well. Protein structure predictions could also contribute to understanding specific diseases with a small number of specialist groups. In the future, unlocking protein shapes could help scientists better understand the natural world and perhaps expand existing knowledge of life itself.
DeepMind has a reputation for being secretive about its work. But when it described AlphaFold 2 in a brief presentation at CASP last year, it promised that it would soon publish the paper outlining the details of the project. David Baker, whose team developed the RoseTTaFold, expressed his frustration at this apparent secrecy of DeepMind. He said, “Among academics, there was a fair amount of doom and gloom. If someone has solved the problem you’re working on but doesn’t disclose how they did it, how do you continue working on it?”
These concerns are put to rest with DeepMind’s decision to open-source AlphaFold 2.0. As per AlphaFold researcher John Jumper, the open-source version is 16 times faster and can generate structures in minutes to hours, depending on protein’s size.
Although the source code is freely available now, it may not be useful for researchers without technical expertise. Pushmeet Kohli, the head of AI for science at DeepMind, has collaborated with select organisations and researchers to predict specific targets and broaden access.
AlphaFold & RoseTTAFold
Inspired by AlphaFold 2.0, a team of researchers from the University of Washington created an alternative open-source model RoseTTAFold. The team claimed that their model achieved similar results as AlphaFold 2.0, under lower computational costs. Soon after, DeepMind published a new paper in Nature magazine giving the details behind the AlphaFold 2.0 model and also open-sourcing the code.
Two ground-breaking algorithms about the same problem in the span of a single week don’t feel like just a coincidence. Besides the speculation, we can’t ignore that some competition has definitely benefited the research community.