TransUNet, a Transformers-based U-Net framework, achieves state-of-the-art performance in medical image segmentation applications. U-Net, the U-shaped convolutional neural network architecture, becomes a standard today with numerous successes in medical image segmentation tasks. U-Net has a symmetric deep encoder-decoder network with skip-connections to improve detail retention. U-Net has excellent representational power but has poor long-range relation due to convolution operations’ intrinsic locality. Medical image segmentation tasks have a common issue of variations in texture, shape, and size of segments. Therefore U-Net with a self-attention mechanism gives a not-bad performance in medical image segmentation. On the other hand, Transformers are considered the alternative architectures to U-Nets that rely only on attention mechanisms. Transformers are good at modelling global contexts by demonstrating superior transferability for downstream tasks under large-scale pretraining. Transformers show exceptional performance in various machine learning tasks, including natural language processing and image recognition. However, transformers concentrate solely on the global context but fail to capture detailed localization information. It results in low-resolution features and, thus, loss of some minute and valuable information.
A lot of extensions or variations of either U-Nets or Transformers are proposed to improve models’ performances. TransUNet has been introduced by Jieneng Chen, Yongyi Lu, Qihang Yu, Xiangde Luo, Ehsan Adeli, Yan Wang, Le Lu, Alan L. Yuille, and Yuyin Zhou as a hybrid-version of U-Net and Transformers by grasping the abilities of both the architectures. TransUNet can capture in-depth localization information at all network stages while holding the power of transferring the context over a long-range within the network. TransUNet yields better results compared to CNN-based self-attention models. It is built as a framework upon Vision Transformer (ViT), the state-of-the-art method on ImageNet classification. TransUNet demonstrates superior as well as state-of-the-art performance in applications such as multi-organ segmentation and cardiac segmentation.
Transformer unit of the framework consists of 12 Transformer layers. A single Transformer layer of the TransUNet framework consists of a stack of a normalization layer, a Multiple Sequence Alignments layers (MSA), a normalization layer and a multi-layer perceptron.
Input images are fed to a CNN unit where the features are extracted at different depth-levels at different layers. This CNN unit acts as the encoder part of the framework. The linear projection of extracted features are fed to the Transformer unit to obtain global context of features. Decoder part of the framework is constructed by feeding the output features of the Transformer unit and skip-connections from the encoder part. The output of the decoder is fed to a Segmentation head where the segmented image version of the original image is obtained.
3-Skip-connections is found to be best among 0, 1, or 3 number of skip connections which yields the highest average dice similarity coefficient (DSC). The best version of the TransUNet framework has 3-skip-connections between the encoder and the decoder parts.
Python Implementation of TransUNet
The public datasets for training or/and testing the framework is available in Google cloud storage in Numpy format. Among various datasets available, we do implementation here using R50+ViT-B_16 dataset. Following are the steps to implement TransUNet in the python environment.
Step-1:
Dataset can be downloaded on the local machine using the following command. A new directory is created to store the dataset so that it can be identified using a pre-trained model.
%%bash wget https://storage.googleapis.com/vit_models/imagenet21k/R50%2BViT-B_16.npz && mkdir -p ../model/vit_checkpoint/imagenet21k && mv R50+ViT-B_16.npz ../model/vit_checkpoint/imagenet21k/R50%2BViT-B_16.npz
Step-2:
Pre-trained TransUNet can be imported on local machine using the command
!git clone https://github.com/Beckschen/TransUNet.git
Output:
The necessary files, python codes for pre-trained weights, training, testing and datasets are downloaded from the corresponding github repository.
Presence of files in the directory can be ensured using the command
%%bash cd TransUNet ls -p
Output:
Step-3:
Necessary libraries, packages can be installed on the local machine by running the requirements.txt file available with the downloaded git files. We have to make sure that the CUDA GPU runtime is enabled. In a notebook environment such as colab or Jupyter, CUDA GPU can be enabled using the Runtime menu option.
%%bash cd TransUNet pip install -r requirements.txt
It may be noted that the current implementation runs on the PyTorch framework.
Step-4:
TransUNet model can be recreated and the weights can be restored back from the pre-trained model using the following command. It can be noted that this command is meant for the R50-ViT-B_16 dataset. For a different dataset, the command can be modified accordingly.
%%bash cd TransUNet/ CUDA_VISIBLE_DEVICES=0 python train.py --dataset Synapse --vit_name R50-ViT-B_16
Based on the device’s memory, the batch size can be reduced to 12 or 6 to save memory. However the ‘base_lr’ variable in the code has to be decreased linearly, and both batch sizes can reach similar performance.
Step-5:
Once trained, the model can be tested with a suitable dataset using the following command. Here testing is done with the Synapse dataset as performed in the original research paper.
%%bash cd TransUNet/ python test.py --dataset Synapse --vit_name R50-ViT-B_16
If the user is interested in testing the model with their own dataset, the data must be saved in the ‘dataset’ directory in Numpy format. 3D images should be clipped within [-125, 275], normalized to [0, 1]. 2D slices should be extracted from 3D volume for training cases while keeping the 3D volume in h5 format for testing cases.
Presence of dataset in the right directory can be confirmed using ‘tree’ command
%%bash sudo apt-get install tree tree
Output:
Performance of TransUNet
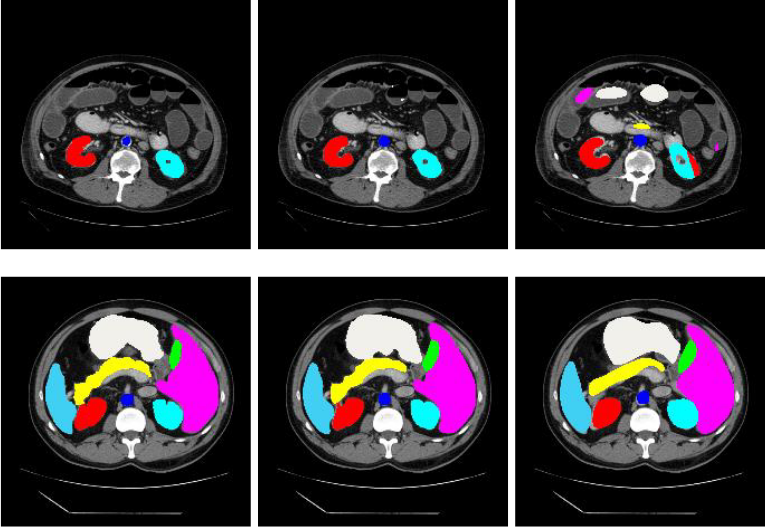
The TransUNet framework is evaluated using the Synapse multi-organ segmentation dataset. This dataset consists of abdominal CT scans with annotations of 8 abdominal organs namely, aorta, gallbladder, left kidney, right kidney, liver, pancreas, spleen, and stomach. Groundtruth images are color-annotated to denote the segments. TransUNet segments the given test image with a relatively greater average dice similarity coefficient (DSC) than any existing state-of-the-art. The average Hausdorff distance (HD), a measure of mis-segmentation, is found minimum in the TransUNet method than other existing methods.
The TransUNet outperforms the existing state-of-the-arts such as Fully Convolutional Neural Networks for Volumetric Medical Image Segmentation (V-Net), Domain Adaptive Relational Reasoning (DARR), Convolutional networks for biomedical image segmentation (U-Net), and Attention Gated Networks (AttnUNet) in terms of average Dice Similarity Coefficient and average Hausdorff distance.
Note: Images other than the code outputs are obtained from the original research paper of TransUNet