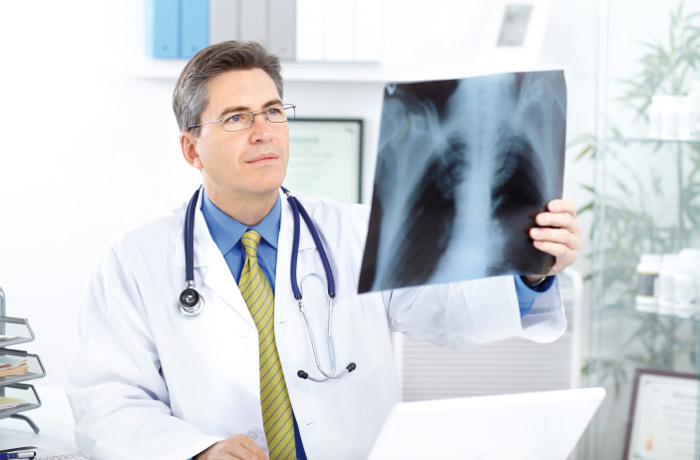
Deep Learning has shown immense results in medical imaging. It is due to the high volume of data that is generated in the medical domain. There are several use cases where AI technologies are used today in the healthcare domain. There can be errors made by humans depending on several factors whereas a machine will not make error provided data is correct. The reason for using Deep learning in medical imaging is the fact that we can attain insights from the data quickly with reliable results. With AI it has now become possible to detect even different types of cancer in the lungs and kidneys also it is used in different therapy. Diagnosing pneumonia is also one of the important applications of deep learning.
Pneumonia is such an infection that is caused in one or both the lungs. This is caused due to viruses, fungi, etc. This results in inflammation in air sacs in the lungs by which it becomes difficult to breathe. Through this article, we will explore how to build a classification model by which we can classify whether a person has pneumonia or not through CXR (Chest X-Ray) images. We will be building the model using pre-trained model Vgg19. For this experiment, we will make use of Pneumonia Chest X Rays data that is publicly available on Kaggle.
The Dataset
There are a total of 5863 CXR (Chest X-Ray) images that are categorized into two categories that are Pneumonia and Normal. The data has three folders: train, test, and Val in which both two categories subfolders are present. The X-rays images were screened by experts so that there are no unreadable images or low-quality images.
The normal chest X-ray (left panel) depicts clear lungs without any areas of abnormal opacification in the image. Bacterial pneumonia (middle) typically exhibits a focal lobar consolidation, in this case in the right upper lobe (white arrows), whereas viral pneumonia (right) manifests with a more diffuse ‘‘interstitial’’ pattern in both lungs. Read more here in this paper.
Model Building
First, we need to install the required package and libraries that are required. As we will be importing the data using the API command from Kaggle. We should have the Kaggle package installed. Use the below code for the same.
from keras.applications.vgg19 import VGG19 from keras.preprocessing.image import ImageDataGenerator from keras.models import Sequential import numpy as np import matplotlib.pyplot as plt import os from zipfile import ZipFile import os from tqdm._tqdm_notebook import tqdm_notebook as tqdm import cv2 import tensorflow as tf import keras !pip install kaggle
Now we will load the dataset from Kaggle and unzip the downloaded file. Refer to the below code to do the same.
from google.colab import files
files.upload()
!mkdir -p ~/.kaggle
!cp kaggle.json ~/.kaggle/
!chmod 600 ~/.kaggle/kaggle.json
!kaggle datasets download -d paultimothymooney/chest-xray-pneumonia
from zipfile import ZipFile
file_name = "chest-xray-pneumonia.zip"
with ZipFile(file_name,'r') as zip:
zip.extractall()
print('Done')
After we have extracted the zip file downloaded from Kaggle we will get three folders that are train, test, and Val. We will now load the training images with the respective labels and visualize normal and pneumonia X-rays. X_train holds the training images and y_train holds the respective labels. Use the below code to do the same.
X_train = []
y_train = []
os.chdir('/content/chest_xray/train/NORMAL')
for i in tqdm(os.listdir()):
img = cv2.imread(i)
img = cv2.resize(img,(256,256))
X_train.append(img)
y_train.append("Normal")
os.chdir('/content/chest_xray/train/PNEUMONIA')
for i in tqdm(os.listdir()):
img = cv2.imread(i)
img = cv2.resize(img,(256,256))
X_train.append(img)
y_train.append("PNEUMONIA")
print(len(X_train))
print(len(y_train))
Output:
There are a total of 5216 images in the training folder. We will now visualize one normal and one pneumonia patient X-ray image. Use the below code for the same
plt.figure(figsize=(5,5))
plt.imshow(X_train[10], cmap="gray")
plt.axis('off')
plt.show()
print(y_train[10])
Output:
plt.figure(figsize=(5,5))
plt.imshow(X_train[4000], cmap="gray")
plt.axis('off')
plt.show()
print(y_train[4000])
Output:
Now we will build the model for classifying the X-rays into the desired two categories. We will be using pre-trained architecture VGG19 and will not train the whole network. We will remove the last layer of the network and will add the last custom layer as we only have 2 classes whereas the model was trained for 1000 classes on the ImageNet dataset. Use the below code to the same.
vgg19 = VGG19(input_shape=[224,224,3], weights='imagenet', include_top=False) for layer in vgg19.layers: layer.trainable = False X = Flatten()(vgg19.output) output = Dense(2, activation='softmax')(X) model = Model(inputs=vgg19.input, outputs=output)
Now we will compile the build model using loss function and optimizer. After compiling we will prepare the training and testing images that can be fed to the model for training. Refer to the below code for the same.
model.compile(loss='categorical_crossentropy',optimizer='adam',metrics=['accuracy']) train_dir = '/content/chest_xray/train' testing_dir = '/content/chest_xray/test' train_datagen = ImageDataGenerator(rescale = 1./255, shear_range = 0.2, zoom_range = 0.2, horizontal_flip = True) test_datagen = ImageDataGenerator(rescale = 1./255) train_data = train_datagen.flow_from_directory(train_dir, target_size = (224, 224), batch_size = 32, class_mode = 'categorical')
test_data = test_datagen.flow_from_directory(testing_dir, target_size = (224, 224), batch_size = 32, class_mode = 'categorical')
history = model.fit(train_data,validation_data=test_data,epochs=10)
Output:
Now we will visualize the model accuracy and model loss for training and testing. Then we will evaluate the model performance. Use the below code to do the same.
plt.plot(history.history['loss'])
plt.plot(history.history['val_loss'])
plt.title('model loss')
plt.ylabel('Loss')
plt.xlabel('Epoch')
plt.legend(['Training', 'Testing'], loc='upper left')
plt.show()
Output:
plt.plot(history.history['accuracy'])
plt.plot(history.history['val_accuracy'])
plt.title('model accuracy')
plt.ylabel('Accuracy')
plt.xlabel('Epoch')
plt.legend(['Training', 'Testing'], loc='upper left')
plt.show()
Output:
model.evaluate(test_data)
Output:
Now we will save the model to compute predictions on random images from the testing data. Use the below code to the same.
model.save('MODEL.h5')
Now we will compute predictions. To do so first we will import the required libraries that are required and load the saved model. Use the below code for the same.
import keras
from keras.models import load_model
import os
from tqdm._tqdm_notebook import tqdm_notebook as tqdm
import cv2
import numpy as np
import matplotlib.pyplot as plt
model = load_model('/content/drive/My Drive/Pneumonia/MODEL.h5')
Let us import the testing data from the test folder.
X_test = []
y_test = []
os.chdir('/content/chest_xray/test/NORMAL')
for i in tqdm(os.listdir()):
img = cv2.imread(i)
img = cv2.resize(img,(224,224))
X_test.append(img)
y_test.append("0")
os.chdir('/content/chest_xray/test/PNEUMONIA')
for i in tqdm(os.listdir()):
img = cv2.imread(i)
img = cv2.resize(img,(224,224))
X_test.append(img)
y_test.append("1")
We will now convert the images into NumPy arrays. After this, we will evaluate the model performance on testing images using different metrics. Use the below code for the same.
X_test = np.array(X_test)
y_test = np.array(y_test)
predicted_classes = model.predict(X_test[:,:,:,:])
predicted_classes = np.argmax(np.round(predicted_classes),axis=1)
predicted_classes[0]
y_test = y_test.astype('int64')
from sklearn.metrics import accuracy_score,classification_report
accuracy_score(predicted_classes,y_test)
Output:
print(classification_report(predicted_classes,y_test))
Output:
Now we will make some predictions on the testing images and compare the results. Use the below code to the same.
L = 3
W = 3
fig, axes = plt.subplots(L, W, figsize = (12,12))
axes = axes.ravel()
print('\n\n\t\t0 Class Represents Normal & 1 Class Represents Pnemonia')
for i in np.arange(0, L * W):
axes[i].imshow(X_test[i])
axes[i].set_title(f"Prediction Class = {predicted_classes[i]}\n True Class = {y_test[i]}")
axes[i].axis('off')
plt.subplots_adjust(wspace=0.5)
Out of 9 testing images 7 images were correctly classified whereas 2 were misclassified by the model.
Conclusion
We have built an AI model using pre-trained architecture VGG19 for classifying X-ray images into pneumonia and normal images. The model classified 7 out of 9 images correctly. The model performance can be more enhanced by getting more data and performing some good no of augmentation techniques referring to the domain knowledge. We can also try making the predictive model using different architectures like Inception or ResNet50 in the same manner and can compare the results. Also, check this article where I classified Brain Tumors from MRI images.